
Principle of cDNA cloning:
- Complementary DNA (cDNA) cloning is termed for the gene cloning (cloning of DNA fragments) obtained from cDNA.
- The principle of cDNA cloning is that it involves the copying of mRNA transcripts into DNA, which are then inserted into bacterial plasmids and then placed into bacteria by transformation.
- At this stage, it should be clear that mRNA used for cDNA preparation is a processed transcript and not the original one transcribed from DNA.
- In-order to clone a DNA sequence that codes for a required gene product, the gene should be removed from the organism and cloned it in the vector molecule.
- A gene library is a random collection of cloned fragments in an appropriate vector that particularly consists of all the genetic information about that species.
- There are two methods for the formation of gene libraries. They are:
- Complementary DNA (cDNA)
- Genomic DNA libraries
Steps of cDNA cloning:
- Isolation of mRNA
- Synthesis of first strand of cDNA
- Synthesis of second strand of cDNA
- Cloning of cDNA
- Introduction to host cells
- Clone selection
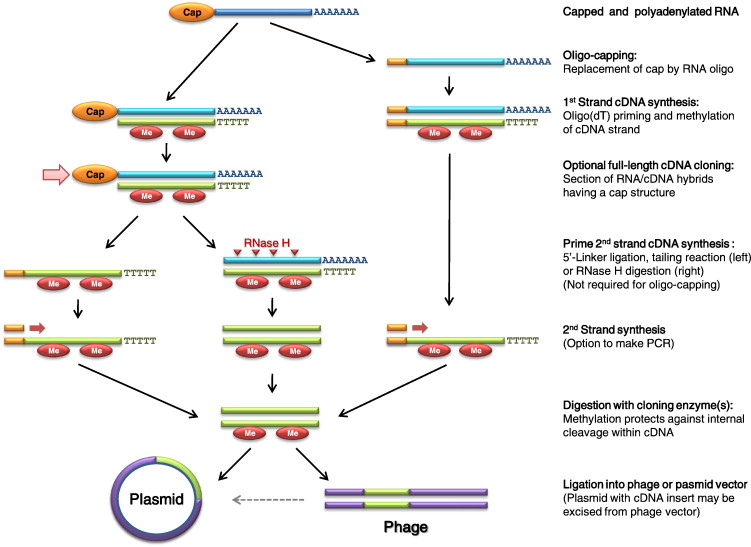
source: sciencedirect.com
1. Isolation of mRNA:
- A crude extract of the tissue with the gene of interest is prepared.
- The extract must be free from proteins, polysaccharides and all other contaminants.
- The technique of oligo-deoxythymine (oligo-dT) cellulose chromatography is used for the further purification of many eukaryotic mRNAs from the total or polysomal fraction.
- mRNAs consist of poly A (adenosine residues) tail at their 3′ end.
- Under favourable conditions, this tail will bind to a string of thymidine residues immobilized on cellulose and then poly (A)+ fraction can be eluted.
- Two or three passages of the poly (A)+ fraction through such a column produces a fraction highly enriched for mRNA.
- This fraction includes different mRNA sequences, however certain techniques can be employed for extracting a particular mRNA species.
- After the preparation of the fraction, it is essential to confirm if the extracted mRNA consists of the sequence of interest.
- It is performed by translation of mRNA in vitro and identification of suitable polypeptides in the products obtained.
2. Synthesis of first strand of cDNA:
- Reverse transcriptase is a RNA dependent DNA polymerase which is used to copy the mRNA fraction into the first strand of DNA.
- This enzyme, like all other DNA polymerases, can only add residues at the 3′-OH group of an existing primer, which is base paired with the template.
- The most commonly used primer is oligo-dT for cloning of cDNAs.
- Oligo-dT primer is 12-18 nucleotides in length, that binds to the poly (A) tract at the 3′ end of mRNA molecules.
- The RNA strand of the resulting RNA-DNA hybrid is destroyed prior to second strand synthesis through alkaline hydrolysis.
3. Synthesis of second strand of cDNA:
- The second strand of cDNA can be synthesized by two techniques. They are:
- i. Self-priming cDNA:
- In Self-priming, the mRNA hybrid obtained is denaturated for the synthesis of second strand on the single strand of cDNA by the klenow fragment of DNA polymerase I.
- The transitory hairpin structure at the 3′ end of single-stranded DNA can be used to prime the synthesis of second strand of cDNA by the klenow fragment of Escherichia coli DNA polymerase I.
- Single-strand specific S1 nuclease digests the hairpin loop and any single-stranded overhung at the other end.
- The ultimate product is a population of double-stranded, blunt-ended DNA molecules complementray to the original mRNA fraction.
- ii. Replacement synthesis:
- In this method, the cDNA:mRNA hybrid works as a template for a nick translation reaction.
- In the mRNA strand of the hybrid, RNase H produces nicks and gaps, creating a series of RNA primers.
- These RNA primers are used by E. coli DNA polymerase I during the synthesis of second strand of cDNA.
- The advantages of this technique are:
- – very efficient
- – can be performed directly using the products of the first strand reaction
- – eliminates the need to use nuclease S1 to cleave the single-stranded hairpin loop in the double stranded cDNA.
4. Cloning of cDNA:
- The most frequently used technique for cloning cDNAs involves the addition of complementary homopolymeric tracts to double stranded cDNA and to the plasmid vector.
- To the cDNA, strings of cytosine residues are added using the enzyme terminal transferase to form oligo-dC tails on the 3′ ends.
- Likewise, a plasmid is cut open at a unique restriction endonuclease site and tailed with oligo-dG.
- Now, the vector and the double stranded cDNA are joined by hydrogen bonding between the complementary homopolymers.
- It results in the formation of open circular hybrid molecules capable of transforming E. coli.
5. Introduction to host cells:
- For the transforming of bacteria, the recombinant plasmids are used, usually the E. coli K-12 strain.
- The uptake of plasmid molecules from the surrounding medium is performed by E. coli cells treated with calcium chloride.
- Any gaps in the recombinant plasmid will be repaired by the host cells.
- The transformed bacteria can be isolated from non-transformed ones on the basis of antibiotic resistance.
- Majority of cloning plasmids contain two antibiotic resistance genes, one of which is destroyed during cloning.
- For instance, in the case of pBR322, cloning into unique PstI site destroys ampicillin resistance but leaves tetracycline resistance intact.
- Bacteria transformed with a recombinant plasmid will be sensitive to ampicillin but resistant to tetracycline.
6. Clone selection:
- The antibiotic resistance selection already performed has recognized which clones carry a recombinant plasmid, however there will be thousands of various inserts.
- The cloning process generally commences with a whole population of mRNA sequences.
- Selection of clones carrying the sequence of interest is the tough job.
- If the gene is expressed, then the simplest selection is to screen for the presence of the protein.
- It can be screened either by bacterial phenotype it produces or by the protein detection methods usually based on immunological or enzymological techniques.
- If the protein is not expressed, then other methods such as nucleic acid hybridization are used.
- Identification of the gene is discussed after the genomic DNA cloning.
