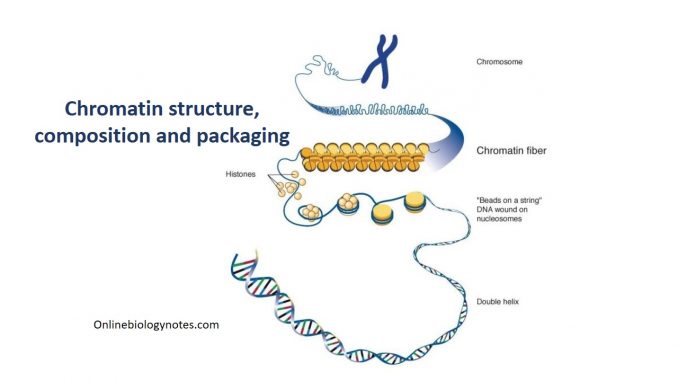
Chromatin:
- Eukaryotic chromosomes are located within a separate cellular compartment termed as nucleus.
- The length of DNA must be compacted by a remarkable amount in order to fit it inside nucleus.
- The compacting of DNA is accomplished by the binding of the DNA to many different cellular proteins.
- The formation of a highly organized DNA-protein complex, termed as chromatin, which is a nucleoprotein complex completes the packing.
- Chromatin is a dynamic structure capable of changing its shape and composition during the life of a cell (cell cycle).
- Chromatin can be defined as highly condensed chromosomes at metaphase stage, and very diffuse structures in course of interphase.
Chromatin composition and packaging
Histones:
- Histones are most abundant proteins in chromatin.
- Histones are small and positively charged proteins and are of 5 major types: H1, H2A, H2B, H3 and H4.
- Histones are characterized by the presence of high percentage of basic amino acids arginine and lysine.
- These amino acids are positively charged that give the histones a net positive charge facilitating the binding of histones to the negatively charged DNA.
- Histone and DNA are present in equal amounts in chromatin.
- A heterogenous variety of non-histone chromosomal proteins also are found in eukaryotic chromosomes.
- There are times where variant histones, with different amino acid sequences, are integrated into chromatin in place of one of the major histone proteins.
- The amino acid sequences of histones H2A, H2B, H3 AND H4 are highly conserved, even between distantly related species.
- Evolutionary conservation of these amino acid sequences highly indicates that histones perform the same basic role in organizing the DNA in the chromosomes of all eukaryotes.
- Structural studies suggest that the histones classes do share a similar tertiary structure, showing that all histones are ultimately evolutionarily related.
Role of H1:
- The next level of condensation of chromatin is brought about by histone H1.
- H1, in contrast to the other histones is not the part of the core particle.
- H1 binds to 20-22 bp of DNA, where the DNA joins and leaves the octamer.
- H1 binds both to the linker DNA at one end of the nucleosome and to the middle of the DNA segment wrapped around core histones.
- H1 serves to restrict the DNA into place and functions as a clamp around the nucleosome octamer.
- The core particle and it’s associated H1 histone are altogether called as the chromatosome.
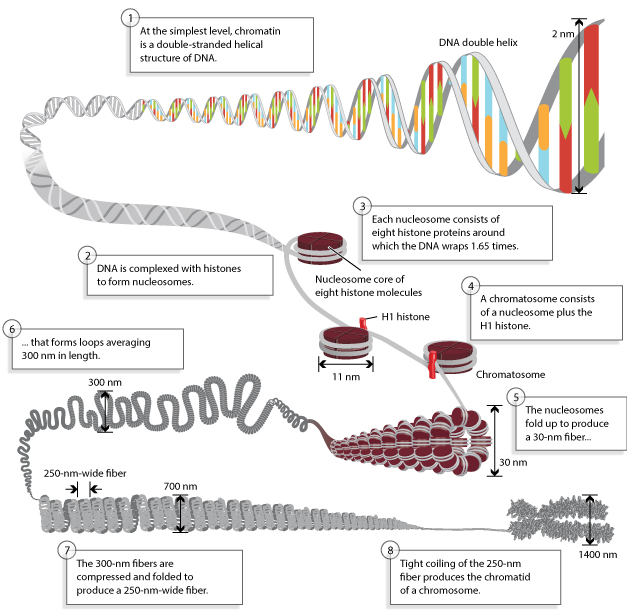
Nucleosomes:
- When the chromatin is isolated from the nucleus of a cell and observed under an electron microscope, it resembles beads on a string.
- The repeating core of protein and DNA produced by digestion with nuclease enzymes is the nucleosome.
- Nucleosome is the basic structural and fundamental unit of chromatin and is the simplest level of chromatin.
- Nucleosome is a core particle formed when the DNA is wrapped about 2 times around an octamer of eight histone proteins (2 copies each of H2A, H2B, H3, H4).
- The DNA in direct contact with the histone octamer is between 145 and 147 bp in length.
- This configuration compacts the DNA by six times.
Linker DNA:
- Each chromatosome encloses about 167 bp of DNA (147bp around nucleosome+20bp bound by H1).
- Chromatosomes are present at regular intervals along the DNA molecule and are apart from each other by linker DNA.
- The size of linker DNA varies among cell types, in most cells, linker DNA comprises of about 30-40 bp.
The 30nm chromatin fiber:
- The nucleosomes compact themselves into a structure about 30nm in diameter, now termed as the 30nm chromatin fiber.
- There are two possible models for the 30nm fiber.
- They are:
- Solenoid model: In this model, a linear array of nucleosomes are coiled into a higher order left handed helix, entitled as solenoid, with around six nucleosomes per turn.
- Helix model: In this model, nucleosomes are arranged in a zigzag ribbon that twists or supercoils.
Higher order structure of chromatin:
- The next higher level of chromatin structure is represented by a series of loops of 30nm fibers, each anchored at its base by proteins in the nuclear scaffold.
- On average, each loop encloses some 20-100kb of DNA and measures about 300nm in length.
- The 300nm loops are packed and folded to result a 250nm wide fiber.
- Tight helical coiling of the 250 nm, in turn, yields the structure that is visible in metaphase- individual chromatids approximately 700nm in width.
Overall, this packaging produces a chromosome that is about 10,000 times shorter and about 400 times thicker, than naked DNA.
