
Introduction of DNA Microarray technique:
- Also termed as DNA chips, gene chips, DNA arrays, gene arrays and biochips.
- Biochips are latest generation of biosensors developed by use of DNA probes.
- DNA microarray is one of the molecular detection techniques which is a collection of microscopic characteristics (commonly DNA) affixed to a solid surface.
- DNA microarrays are solid supports usually made up of glass or silicon upon which DNA is attached in an organized pre-arranged grid design.
- Each spot of DNA, termed as probe, signifies a single gene.
- DNA microarrays can examine the expression of tens of thousands of genes concurrently.
- There are 2 types of DNA microarray i.e. cDNA based microarray and oligonucleotide based microarray.
Principle of DNA microarray:
- DNA microarray technology was originated from Southern blotting, in which fragmented DNA is attached to a substrate and then probed with a known DNA sequence.
- DNA microarray is based on principle of hybridization between the nucleic acid strands.
- Complementary nucleic acid sequences have the characteristic to specifically pair to each other by the formation of hydrogen bonds between complementary nucleotide base pairs.
- Unknown sample of DNA sequence is termed as sample or target and the known sequence of DNA molecule is called as probe.
- Fluorescent dyes are used for labelling the samples and at least 2 samples are hybridized to the chip.
- A large number of complementary base pairs in nucleotide sequence is suggestive of tighter non-covalent bonding between the two strands.
- Following the washing off of non-specific bonding sequences, only strongly paired strands will stay hybridized.
- Thus, the fluorescent labeled target sequences that pairs to the probe releases a signal that relies on the strength of the hybridization detected by the number of paired bases, hybridization conditions, and washing after hybridization.
- DNA microarrays employs relative quantization in which the comparison of same character is done under two different conditions and the identification of that character is known by its position.
- After completion of the hybridization, the surface of chip can be examined both qualitatively and quantitatively by use of autoradiography, laser scanning, fluorescence detection device, enzyme detection system.
- The presence of one genomic or cDNA sequence in 1,00,000 or more can be screened in a single hybridization by using DNA microarray.
Types of DNA microarray:
- cDNA based microarrays
- Oligonucleotide based microarrays
cDNA based microarrays:
- cDNA is used for the preparation of chips.
- cDNAs are amplified by PCR.
- It is a high throughput technique.
- It is highly parallel RNA expression assay technique that allows quantitative analysis of RNAs transcribed from both known and unknown genes.
Oligonucleotide based microarrays:
- In this type, the spotted probes contains of short, chemically synthesized sequences, 20-25 mers/gene.
- Shorter probe lengths allows less errors during probe synthesis and enables the interrogation of small genomic regions, plus polymorphisms
- Despite being easier to produce than dsDNA probes, oligonucleotide probes need to be carefully designed so that all probes acquire similar melting temperatures (within 50 c) and eliminate palindromic sequences.
- The probe’s attachment to the glass slides takes place by the covalent linkage as electrostatic immobilization and cross-linking can result in significant loss of probes during wash steps due to their small size.
- The coupling of probes to the microarray surface takes place via modified 5′ to 3′ ends on coated slides that provide functional groups (epoxy or aldehyde)
Requirements of DNA microarray:
- DNA chip
- Fluorescent dyes
- Fluorescent labelled target/sample
- Probes
- Scanner
Steps involved in cDNA based microarray:
- Sample collection
- Isolation of mRNA
- Creation of labeled cDNA
- Hybridization
- Collection and analysis
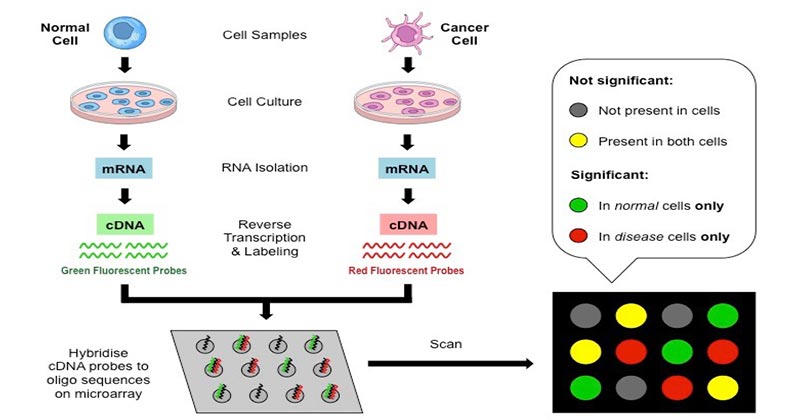
Sample collection:
- A sample can be any cell/tissue that we desire to conduct our study on.
- Generally, 2 types of samples are collected, i.e. healthy and infected cells, for comparing and obtaining the results.
Isolation of mRNA:
- The extraction of RNA from a sample is performed by using a column or solvent like phenol-chloroform.
- mRNA is isolated from the extracted RNA leaving behind rRNA and tRNA.
- As mRNA has a poly-A tail, column beads with poly-T tails are employed to bind mRNA.
- Following the extraction, buffer is used to rinse the column inorder to isolate mRNA from the beads.
Creation of labeled cDNA:
- Reverse transcription of mRNA yields cDNA.
- Both the samples are then integrated with different fluorescent dyes for the production of fluorescent cDNA strands which allows to differentiate the sample category of the cDNAs.
Hybridization:
- The labeled cDNAs from both the samples are placed on the DNA microarray which permits the hybridization of each cDNA to its complementary strand.
- Then they are thoroughly washed to remove unpaired sequences.
Collection and analysis:
- Microarray scanner is used to collect the data.
- The scanner contains a laser, a computer and a camera. The laser is responsible for exciting the fluorescence of the cDNA, generating signals.
- The camera records the images produced at the time laser scans the array.
- Then computer stores the data and yields results instantly. The data are now analyzed.
- The distinct intensity of the colors for each spot determines the character of the gene in that particular spot.
Applications of DNA microarray technique:
- Drug discovery
- Study of functional genomics
- DNA sequencing
- Gene expression profiling
- Study of proteomics
- Diagnostics and genetic engineering
- Toxicological researches
- Pharmacogenomics and theranostics
