There are many different types of transposons in eukaryotes which vary in size, structure, composition and behavior. All the eukaryotic transposons have basic characteristics such as- having terminal inverted repeats, and target specific duplication when inserted.
Types of transposons in eukaryotes:
I. Class1: Retro-transposons:
- Examples: Ty elements in yeast, copia elements in Drosophila, LINEs and SINEs sequences in humans.
II. Class2: DNA transposons:
- Example: P element in Drosophila
- These are cut and paste transposons; similar to bacterial transposons
I. Class 1: Retro-transposons
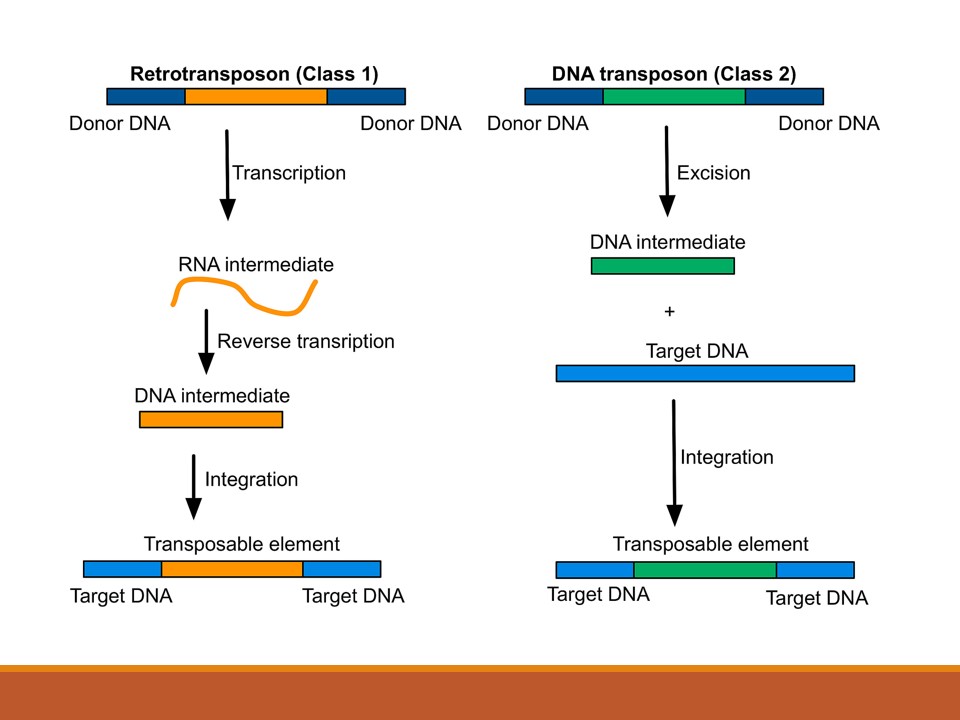
- Retro-transposons are those transposable elements that utilize reverse transcriptase enzyme to convert RNA intermediate into DNA and then transpose into new position by itself.
- They are of two types on the basis of Long terminal repeats (LTR)
1. LTR retro-transposons or retrovirus like element:
- All LTR retro-transposons have the same basic structure: a central coding region flanked by long terminal repeats (LTR) which are oriented in the same direction.
- The LTR are typically a few hundred nucleotide pairs long sequence having short inverted repeats at both ends like present in other types of transposons.
- Due to the characteristic LTR sequence which is similar to that of retrovirus, these transposons are known as LTR retrotransposons.
- The property of LTR retroposons is similar to retrovirus infection.
- LTR retrotransposons utilizes reverse transcriptase enzyme from retro virus that convert transposons RNA into DNA in the first stage and in in second stage the synthesized DNA is transposed into new location in the chromosome.
- Examples: Ty elements of yeast, and copia elements of Drosophila
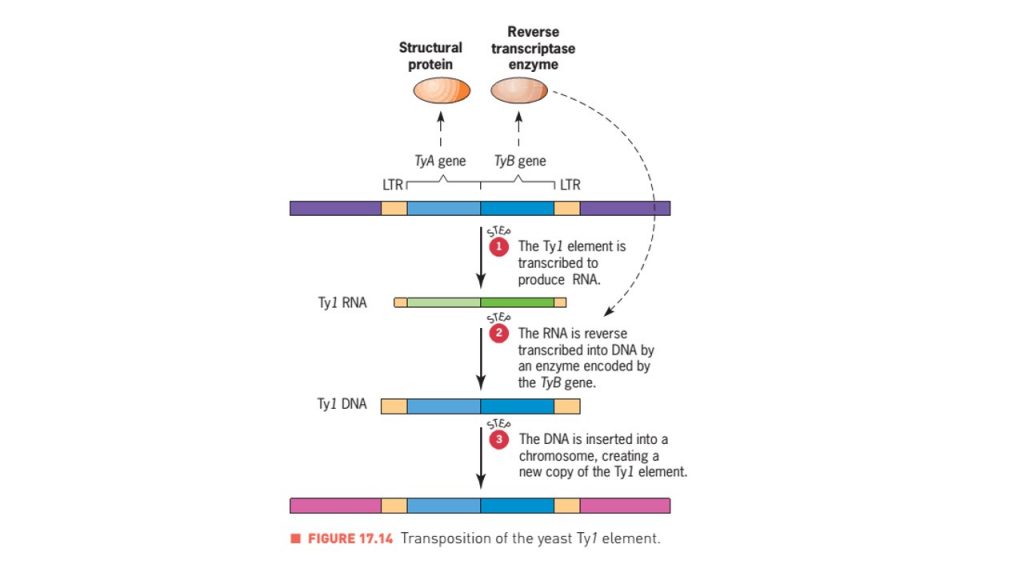
Ty element in yeast
- Ty element is a LTR retro-transposons present in yeasts which is about 5.9 kbp long sequence having LTRs of about 340 base pairs long at both end.
- Most yeast strains have about 35 copies of the Ty1 element in their genome.
- Ty1 elements have only two genes, TyA and TyB, which are homologous to the gag and pol genes of the retroviruses.
- Transposition of Ty1 element involves reverse transcriptase enzyme.
- After RNA is synthesized from Ty1 DNA, a reverse transcriptase encoded by the TyB gene uses it as a template to make double stranded DNA.
- Then the newly synthesized DNA is transported to the nucleus and inserted in targeted site in the genome, creating a new Ty1 element
2. Non-LTR retrotransposons (retroposons):
- The non-LTR retrotransposons are common retrotransposons present in eukaryotes.
- They are simply known as retroposons or non viral transposons
- Non-LTR retrotransposons lacks long terminal repeats (LTRs) at the terminal end. However, they have homogeneous sequence of A:T base pairs at one end which is derived from post transcriptional modification (poly A tail).
- These non-LTR retrotransposons are transposed via RNA intermediate which is reversed transcribed into double stranded DNA by reverse transcriptase enzyme encoded by the element itself and then transposed to the targeted site.
- Examples: Transposable element of human
- LINEs (long interspersed nuclear elements)
- SINEs (short interspersed nuclear elements)
Transposable element of human:
- At least 44 percent of human genome is repetitive which is derived from transposable elements, including retroviruslike elements (8%), retroposons (33 %), and several families of elements that transpose by a cut-and-paste mechanism (3 %)
- Class of human transposons:
- i. LINEs
- ii. SINEs
Long Interspersed nuclear elements (LINEs):
- L1 retroposon is the principal human transposable element which belongs to long interspersed nuclear elements (LINEs) class.
- Two other LINEs sequences, L2 and L3 LINEs are also present in human genome, however both are transpositionally inactive.
- Complete L1 element is about 6 kb long sequence with an internal promoter that is recognized by RNA polymerase II, and two open reading frames (ORFs);
- ORF1 encodes a nucleic acid-binding protein,
- ORF2 encodes a protein with endonuclease and reverse transcriptase activities.
- The human genome contains about 3000 to 5000 copies of complete L1elements.
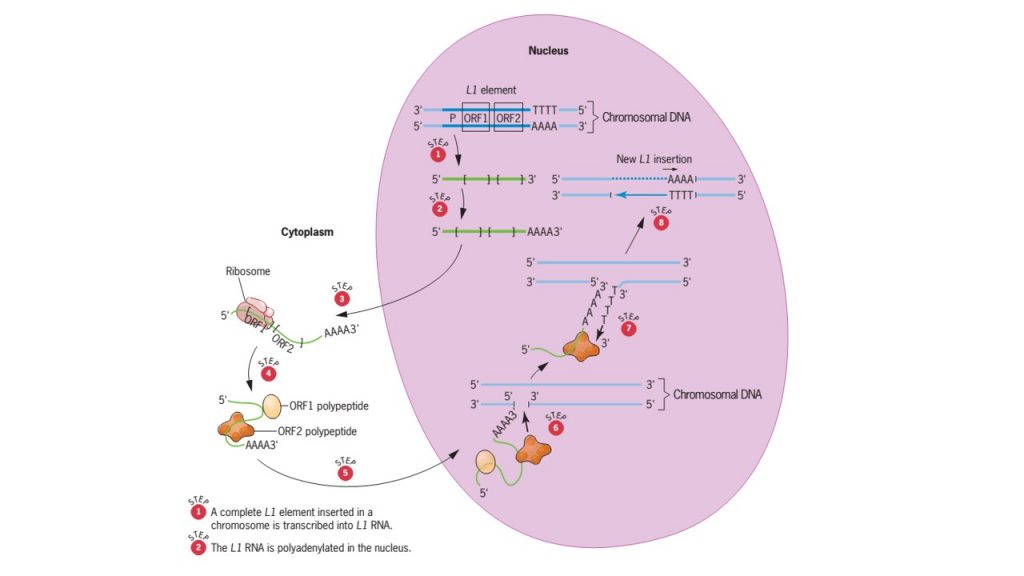
- Transposition of L1 element involves transcription of a complete L1 element into RNA in the first step and the reverse transcription of this RNA into DNA in next step.
- Both transcription and reverse transcription processes take place in the nucleus. However, before the L1 RNA is reverse transcribed, it travels to the cytoplasm where it is translated into polypeptides (nucleic acid binding proteins, endonuclease and reverse transcriptase).
- The polypeptides remain attached with RNA when it returns to the nucleus.
- The polypeptide encoded by ORF2 possesses an endonuclease activities that catalyzes cleavage of one strand of the DNA duplex at targeted site in a chromosome.
- The polyA tail of polypeptide juxtaposed with 5’end of cleaved DNA and the reverse transcriptase activity results into synthesis of single DNA strand.
- Utilizing 3’ end as a primer for DNA synthesis, the newly synthesized L1 DNA is subsequently made double-stranded. Therefore, new L1 element is placed in new location.
Short interspersed nuclear elements (SINEs):
- The short interspersed nuclear elements (SINES), are the second most abundant class of transposable elements in the human genome.
- SINEs are less than 400 base pairs long and have an internal promoter but do not encode proteins.
- Like all retroposons, SINEs do not have terminal repeats but instead they have a sequence of A:T base pairs at one end.
- Transposition of SINEs involves transcription of a SINEs DNA into RNA which is then reversed transcribed into DNA.
- However, SINEs do not have any open reading frame to encode any enzymes. Therefore, it depends upon LINEs-type element for reverse transcriptase enzyme and other enzymes.
- Thus, the SINEs in depend on the LINEs to multiply and insert within the genome.
- Three families of SINEs are present in Human genome- Alu, MIR, and Ther2/MIR3 elements.
- However, only the Alu elements is transpositionally active
II. Class 2: DNA Transposons
- Some mobile elements found in eukaryotes are DNA transposons and the mechanism of their transposition is similar to those of bacteria.
- The first transposable elements discovered by McClintock in maize are now known to be DNA transposons. However, the first DNA transposons to be molecularly characterized were the P elements in Drosophila.
P element in Drosophila:
- P element present in Drosophila resembles to Insertion sequence (IS element) of bacteria, which possesses a short inverted terminal repeats at both end and a single open reading frame which encode single protein (transposase).
- The P elements vary in size, ranging from 0.5 to 2.9 kb in length.
- P elements were discovered by Margaret Kidwell, who was studying hybrid dysgenesis
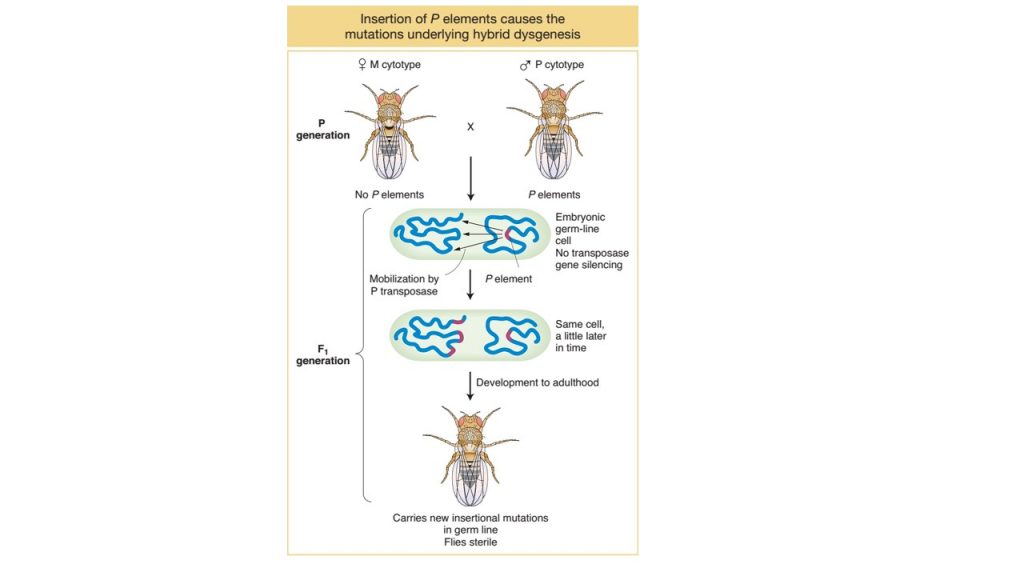
Dysgenesis in Drosophila:
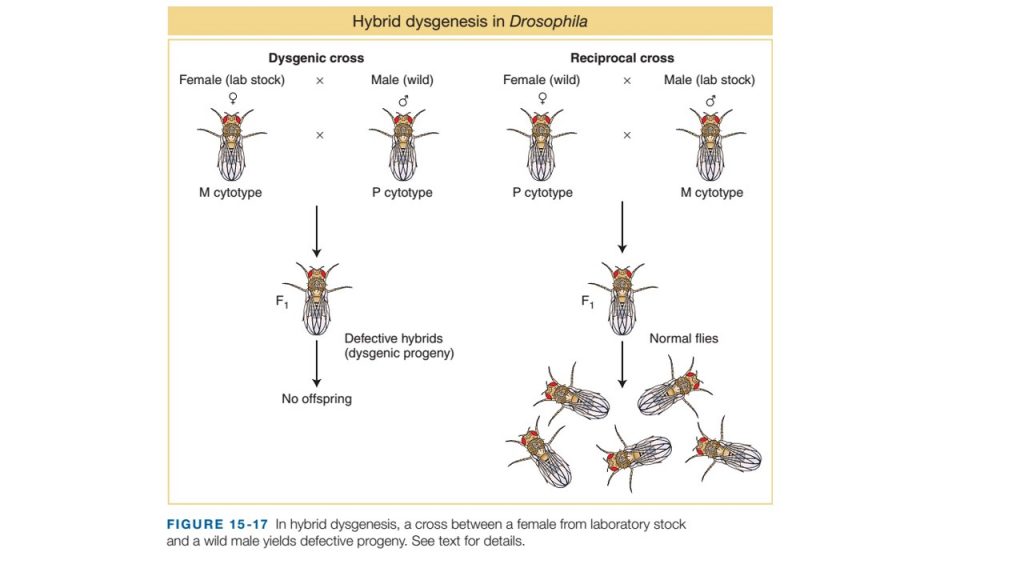
- A phenomenon known as dysgenesis is present in Drosophila when laboratory reared female flies were crossed with wild male flies.
- In such crosses, the laboratory stocks are said to possess an M cytotype (cell type), and the natural stocks are said to possess a P cytotype. Laboratory flies do not have P element.
- In such cross of M (female) ×P (male), the F1 progeny show a range of surprising phenotypes characters in germ line including sterility, a high mutation rate, and a high frequency of chromosomal aberration and nondisjunction.
- These F1 hybrid progeny are biologically deficient and are known as dysgenic. The phenomenon is called dysgenesis
- However in reciprocal cross between P (female) × M (male), no dysgenesis is observed in F1 progeny.
- To explain the phenomenon of dysgenesis, the simple answer is that P-element transposition. In all adult P cytotype flies, they possess P-element which contains a transposase genes are silenced. However, the genes are activated in the F1generation resulting in dysgenesis when crossed M (female) with P (male)
- In the cross between M cytotype (female flies, no P elements) × P cytotype (male flies, P elements), each parent contribute respective gametes with respective cytotype. Male gamete provide P elements in the newly formed zygote which is in a silencing-free environment, since female gamete is M cytotype. The P elements derived from the male genome can now transpose throughout the zygote genome, causing a variety of damage as they insert into genes and all F1 progeny developed from such zygote are dysgenic.
- On the other hand, in reciprocal cross between P cytotype (female flies, P element) ×M cytotype (male flies, no P-element) no dysgenesis is observed. It is because, in this case female gamete provide P element in the newly formed zygote but the P-element is already silenced in female gamete due to presence of some component in cytoplasm of female gamate. Therefore Zygote developed from such gamete with already silenced P element prevent dysgenesis.