
There are two main type of transposable elements in bacteria having different size and structure. They are;
- Insertion sequences (IS elements)
- Prokaryotic Transposons (Tn): Composite and non-composite transposons
1. Insertion sequences (IS element):
- IS elements are the simplest type of bacterial transposable sequences that can insert at different location of bacterial chromosome and plasmid through illegitimate recombination.
- They are typically short sequences and contains only one gene that encode the enzyme for transposition.
- IS elements were first identified as spontaneous insertion in certain lac operon mutations of E. coli which inactivate the gene and inhibit transcription and translation. The mutation of Lac operon gene was found to be unstable and molecular analysis reveals the presence of extra copies of DNA sequences near the lac gene. When the mutated E. coli undergoes reverse mutation, the extra DNA sequence is lost.
- A bacterial chromosome may contain several copies of a particular type of IS element. For example, 6 to 10 copies of IS1 are found in the E. coli chromosome.
- Plasmids may also contain IS elements
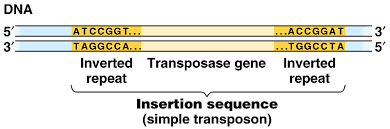
Characteristics of Insertion sequences (IS element):
- IS elements are compactly organized and containing about 1000 nucleotide pairs and contain only genes (open reading frame) which encode for enzyme for regulating transposition.
- Many distinct types of IS elements have been identified. The smallest IS element is IS1 which is 768 nucleotide pairs long.
- Each type of IS element contains inverted terminal repeats at both end and a transposon sequence in between those inverted repeats. Transposon is the only gene that code for transposition of IS element.
- The inverted terminal repeats is 9-40 base pair long and is the characteristics of most IS element
- IS element have the capacity to duplicate the inserted sequence at the site of insertion; known as target site duplication.
Transposition of insertion sequence in bacteria:
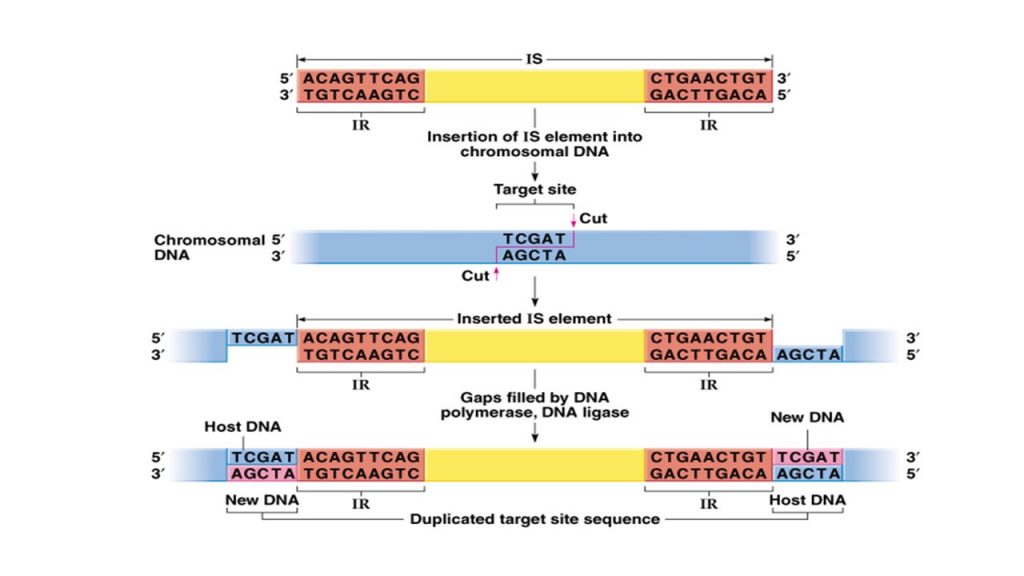
- IS element contains single open reading frame (ORF) which encodes for the enzyme transposase, catalyzing its own transposition.
- The enzyme transposase is like restriction endonuclease which binds to terminal inverted repeats (IR) of IS element which is the restriction site. Then the enzyme cut and excise IS elements from chromosome or plasmid.
- The excised IS element is mobile in nature and moves along the length of chromosome to recognizes the target site for insertion on same or different chromosome or plasmid.
- Once recognizing the target site, it generate staggered cleavage (cut the single strand of DNA) generating sticky and itself get inserted.
- As IS element get inserted, the proofreading mechanism of DNA results in duplication of the DNA sequence at the target site of the insertion such that one copy of target DNA is located on each side of IS element.
- Thus IS elements helps in target site duplication
2. Prokaryotic Transposons (Tn):
- Prokaryotic Transposons are similar to IS element but they are larger and also contains other genes (mostly antibiotic resistance gene) in addition to gene that encode transposase.
- Transposons are several thousand base pairs long and contains inverted terminal repeats.
- There are two types of prokaryotic transposons- composite and non-composite transposons.
- The composite transposons and Tn3-like elements are more complex than IS elements, containing some genes that encode products unrelated to the transposition process.
i. Composite transposons:
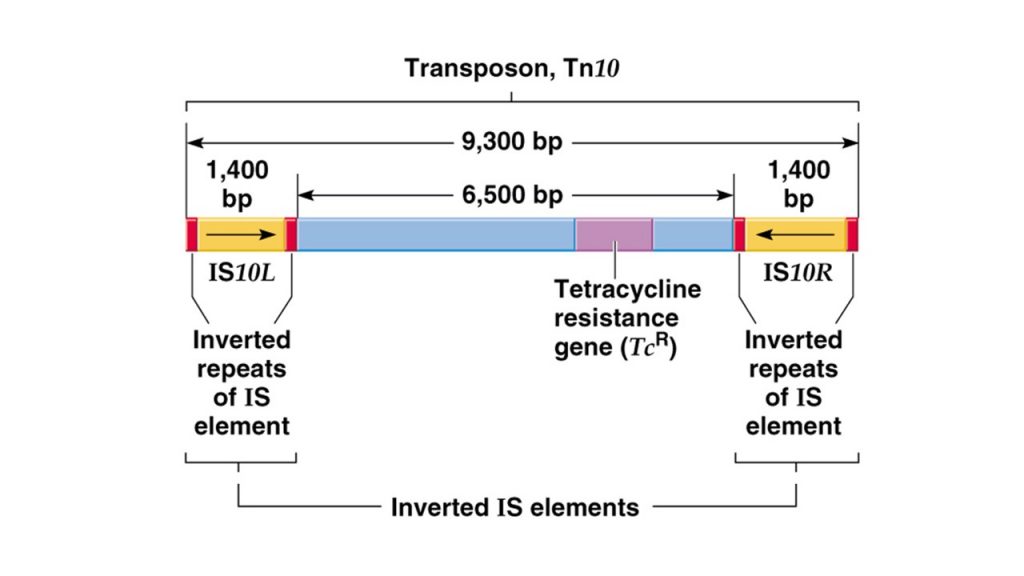
- Composite transposon are created when two IS elements insert near each other and the region between the two IS elements can then be transposed when the elements act jointly.
- For example: Tn10 is a composite transposons of 9.3kbp which contains 1.4 kbp terminal inverted repeats and in between them is gene for transposase and gene for antibiotic resistance.
ii. Non-composite transposons:
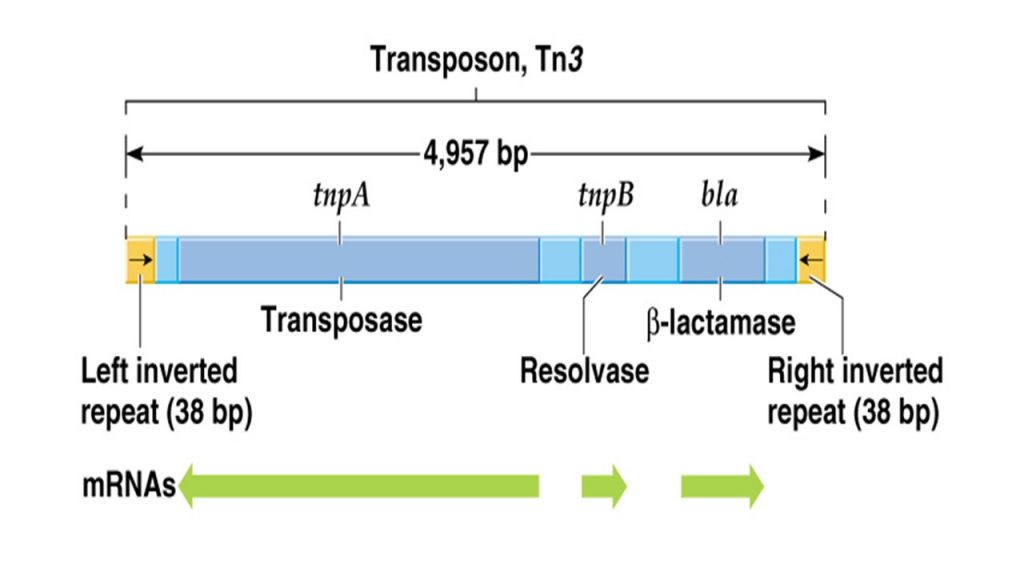
- The non-composite transposons is a sequence of DNA containing gene for trasnposase and multiple other gene in between terminal inverted repeats.
- Unlike composite transposons, it does not contains IS elements at each end but instead it contains simple inverted repeats 0f 38-40 nucleotide pairs at each end.
- For example; Tn3 is a non-composite transposons of 5kbp which contains three gene for beta-lactamase (bla), transposase (tnpA) and resolvase (tnpB).
- The beta lactamase provide resistance to the antibiotic ampicillin, and the other two enzymes play important roles in transposition and recombination.
Role of bacterial transposons in antibiotic resistance:
- As many bacterial transposons also carry genes for antibiotic resistance apart from gene for transposase and by the nature of transposable element, the genes move or translocate from one DNA to another and from chromosome to plasmid and vice versa. This results in genetic flux of antibiotic resistance genes in bacterial population.
- When the transposons undergoes recombination with plasmid vector within a bacterial cell then it can be transformed horizontally or vertically to other bacteria, spreading the drug resistance gene in bacterial population. This creates multidrug resistance pathogenic bacteria such that diseases become difficult to control.
