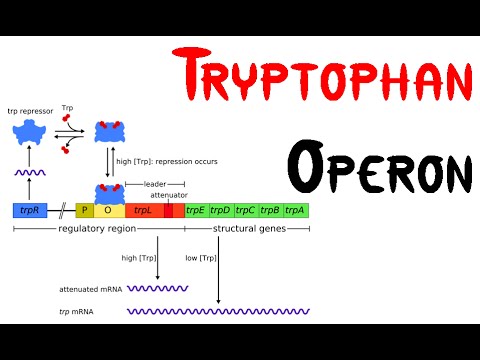
Tryptophan operon
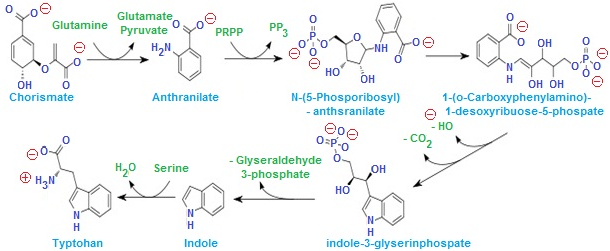
- The tryptophan operon is the regulation of transcription of the gene responsible for biosynthesis of tryptophan from chorismate.
Tryptophan operon consists of structural gene and regulatory gene.
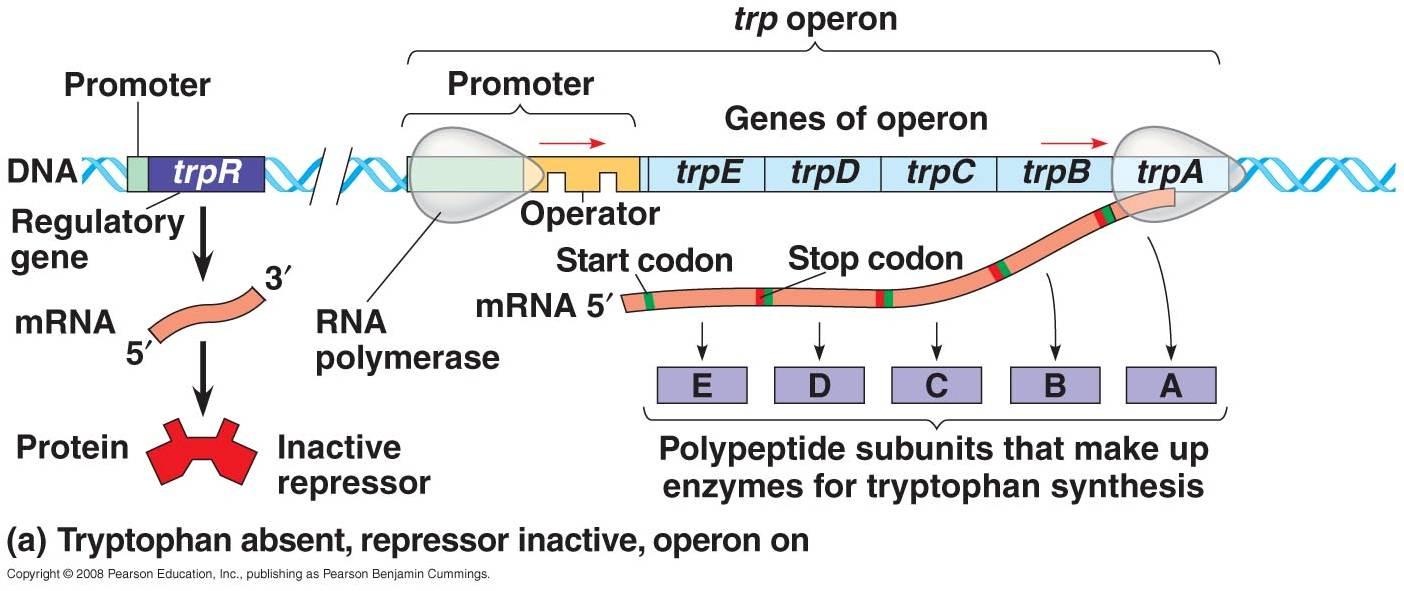
I. Regulatory gene are Promoter, Repressor, Operater and Leader sequence.
II. Structural gene are TrpE, TrpD, TrpC, TrpB and TrpA
- trpE: It enodes the enzyme Anthranilate synthase I
- trpD: It encodes the enzyme Anthranilate synthase II
- trpC: It encodes the enzyme N-5’-Phosphoribosyl anthranilate isomerase and Indole-3-glycerolphosphate synthase
- trpB: It encodes the enzyme tryptophan synthase-B sub unit
- trpA: It encode the enzyme tryptophan synthase-A sub unit
Tryptophan operon is regulated by following mechanism
1. Repression
i. when tryptophan is absent in cell:
- Repressor gene (trpR) encodes the repressor proten which is originally inactive. In the absence of tryptophan, transcription of structural gene occur for the biosynthesis of tryptophan from chorismate.
ii. when tryptophan is high in cell: 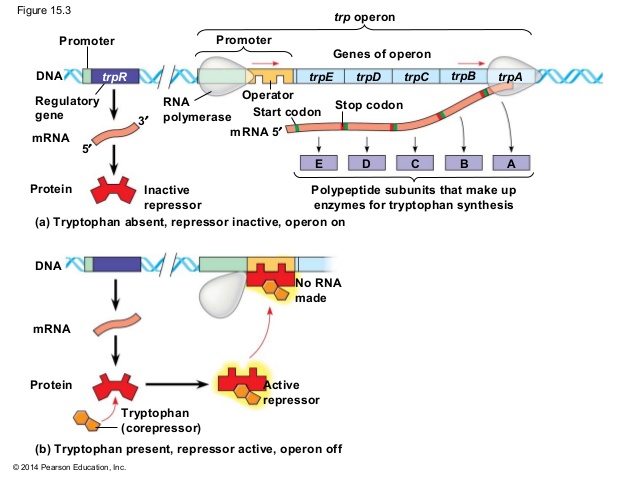
- When tryptophan is high in cell then it binds with repressor protein and change its confirmation so that it become active and bind to the operator near promoter.
- Binding of repressor protein to operator overlaps the promoter, so RNA polymerase cannot bind to the prometer. Hence transcription is halted.
- Since tryptophan is already high in cell, no transcription of structural gene is required for biosynthesis of tryptophan. This is also known as negative regulation.
2. Attenuation:
- In bacteria, transcription and translation occurs simultaneously. The translation starts before transcription completes. In this attenuation mechanism, rate of translation determines whether transcription continues or terminates. Therefore the attenuation mechanism is only found in bacteria but not in eukaryotic cell.
- Leader sequence (trpL) play important role in attenuation. Leader sequence contains such a nucleotide sequence that mRNA transcribed from it contains four specific region. Region 1, region 2, region 3 and Region 4. Region 3 is complementary to both region 2 and region 4.
- If region 3 and region 4 base pair with each other, they form a loop like structure called attenuator and it function as transcriptional termination. If pairing occur between region 3 and region 2, then no such attenuator form so that transcription continues.
- Region 1 is the most important region that determines whether to form loop between region 2-region 3 or region 3-region 4. The region 1 consists of sequence of 14 codons, out of which two codons are tryptophan codon (codon 10 and 11).
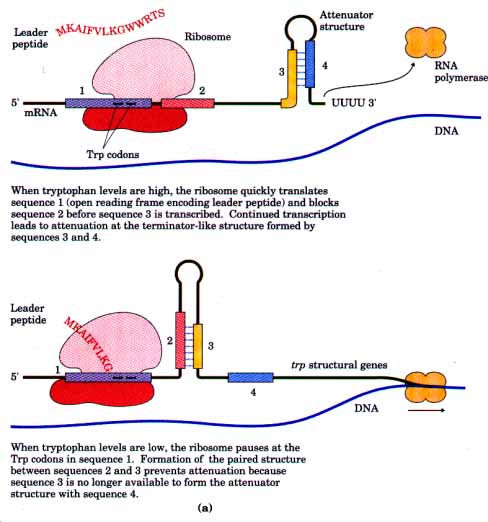
- When tryptophan is high in cell than tRNA crrying tryptophan encodes codon 10 and 11. Such that ribosome encloses the region 2 which is near to the tryptophan codon. Hence region 3 base pair with region 4 to form attenuator as region 2 is not available for pairing. Consequently, transcription is halted.
- When tryptophan is low or absent in cell, then translation stops at the position of tryptophan codon. Such that loop between region 2 and region 3 forms. Transcription continues.
Feedback mechanism of trp operon
- When tryptophan is high in cell then transcription of structural gene does not occur.
- When tryptophan is absent or very low then transcription continues
- If charismate is high in cell then it favour the transcription of structural gene
- If charismate is low or absent in cell then it inhibit transcription
